
Minimising the Side-chains
Once the initial conformations had been proposed we then tried to minimise the energy of each site using the CHARMm forcefield within QUANTA. The minimisations were performed one at a time with the whole protein fixed and only the two (or in the case of site 10, three) residues and the magnesium atom allowed to alter their configuration. The following two graphics show the protein fitted with the electrostatic surface from two different angles to make all (except site 10) visible. To look at the details for each site, click on the site label or scroll down the page. For each of the magnified sidechain diagrams, the minimised structure is shown in blue and the original in green. The Magnesiums are also in blue.
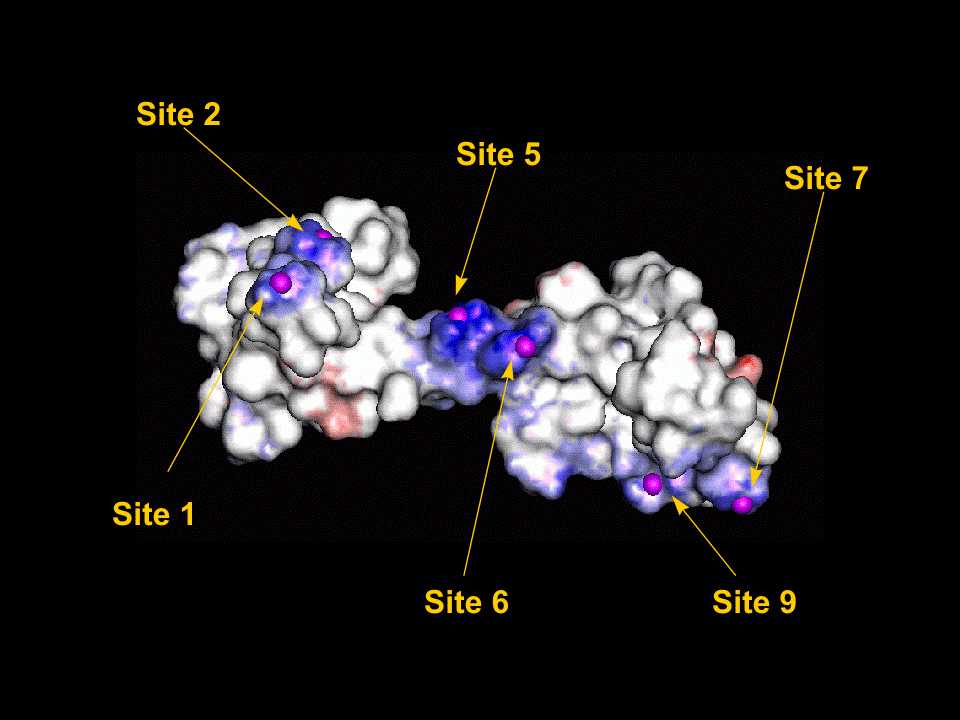
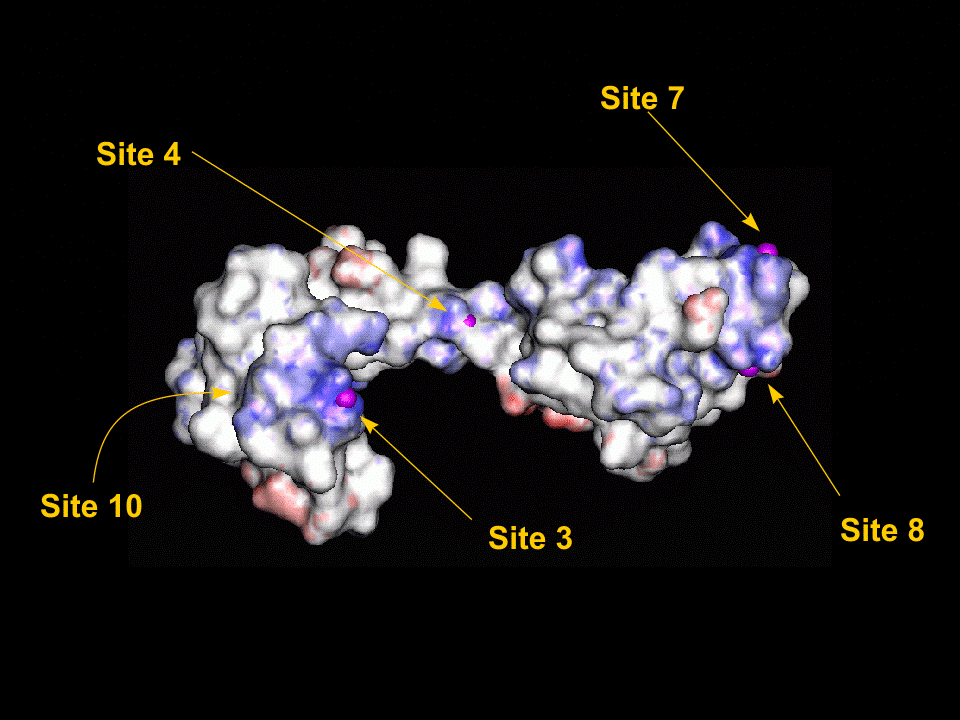
Site 1
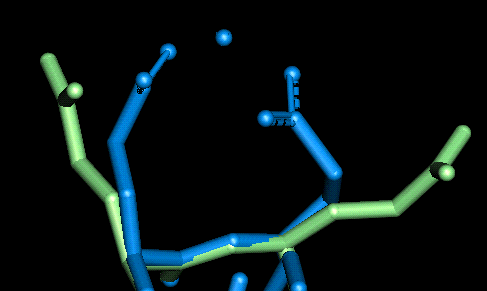
| This site involves Glu 6 and Glu 7. The O - Mg distances in Angstroms are as follows:
- Glu 6: 2.097, 2.071 (bi-dentate)
- Glu 7: 2.101, 2.135 (bi-dentate)
|
Site 2
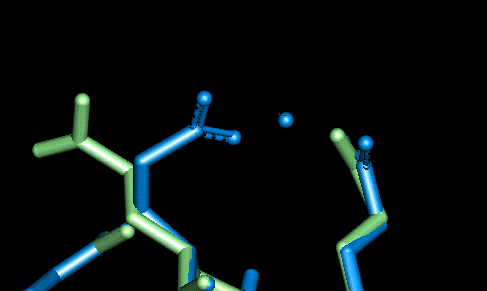
| This site involves Glu 11 and Glu 14. The O - Mg distances in Angstroms are as follows:
- Glu 11: 2.073, 2.172 (bi-dentate)
- Glu 14: 2.164, 2.092 (bi-dentate)
|
Site 3
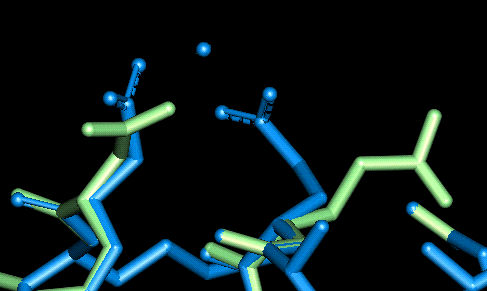
| This site uses Glu 47 and Asp 50. The O - Mg distances in Angstroms are as follows:
- Glu 47: 2.059, 2.101 (bi-dentate)
- Asp 50: 2.022, 3.940 (mono-dentate)
|
Site 4
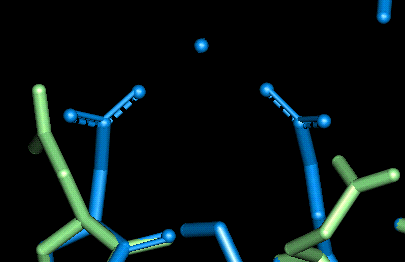
| This site uses Glu 78 and Asp 82. The O - Mg distances in Angstroms are as follows:
- Glu 78: 2.141, 2.101 (mono-dentate)
- Asp 82: 3.153, 5.337 (mono-dentate)
|
Site 5
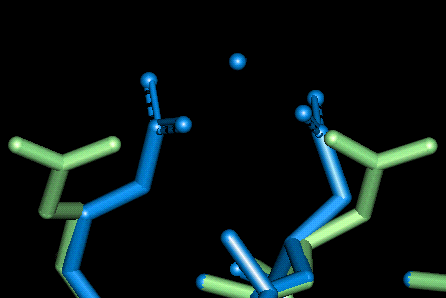
| This site uses Asp 80 and Asp 83. The O - Mg distances in Angstroms are as follows:
- Asp 80: 2.178, 2.115 (bi-dentate)
- Glu 83: 2.143, 2.104 (bi-dentate)
|
Site 6
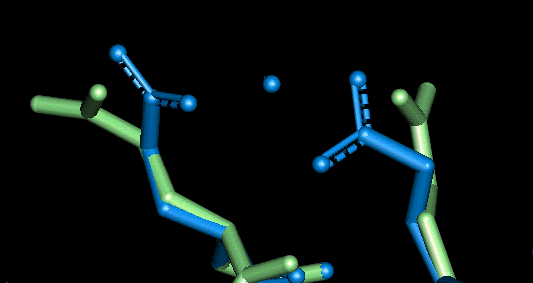
| This site uses Glu 84 and Glu 87. The O - Mg distances in Angstroms are as follows:
- Glu 84: 2.015, 3.843 (mono-dentate)
- Glu 87: 2.058, 2.067 (bi-dentate)
|
Site 7
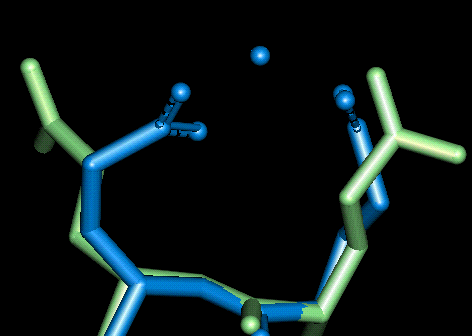
| This site uses Glu 119 and Glu 120. The O - Mg distances in Angstroms are as follows:
- Glu 119: 2.092, 2.114 (bi-dentate)
- Glu 120: 2.065, 2.061 (bi-dentate)
|
Site 8
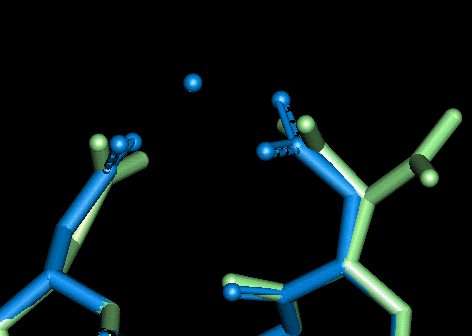
| This site uses Asp 118 and Asp 122. The O - Mg distances in Angstroms are as follows:
- Asp 118: 2.149, 2.121 (bi-dentate)
- Asp 122: 2.107, 2.129 (bi-dentate)
NB The unminimised conformation (in green) had two different positions for the sidechain in the original (1cll.pdb) data file. This was attributed to disorder in the pdb but one of the conformation was positioned almost perfectly for small cation binding.
|
Site 9
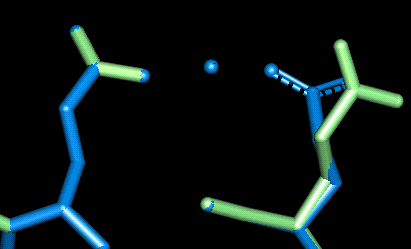
| This site uses Glu 123 and Glu 127. The O - Mg distances in Angstroms are as follows:
- Glu 123: 1.597, 3.595 (mono-dentate)
- Glu 127: 1.900, 3.615 (mono-dentate)
NB The side-chains for these residues would for some reason not sensibly be minimised and minimisation resluted in highly anomolous atom movement.
|
Site 10
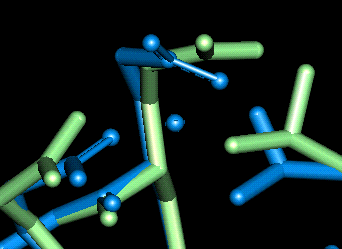
| This site uses Thr 29, Glu 45 and Gln 49. The O - Mg distances in Angstroms are as follows:
- Thr 29: 2.177
- Glu 45: 2.070, 2.585 (bi-dentate)
- Gln 49: 2.284
|
Introduction
Finding a Structure
Sidechain Manipulation
Minimisiation
Conclusions
E-mail me here at barton@yorvic.york.ac.uk













